Reprogramming of macrophages employing gene regulatory and metabolic network models | PLOS Computational Biology

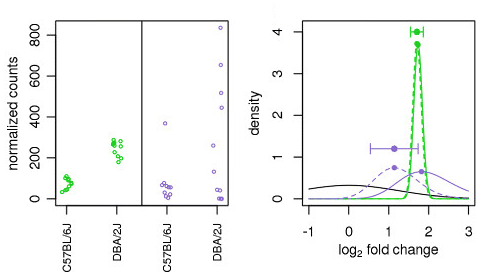
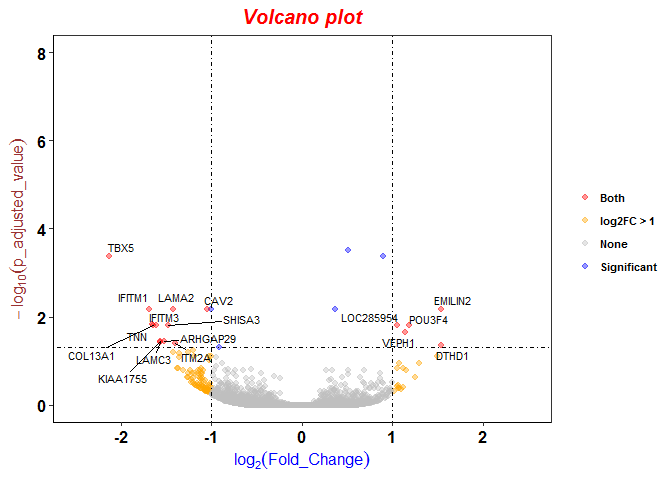
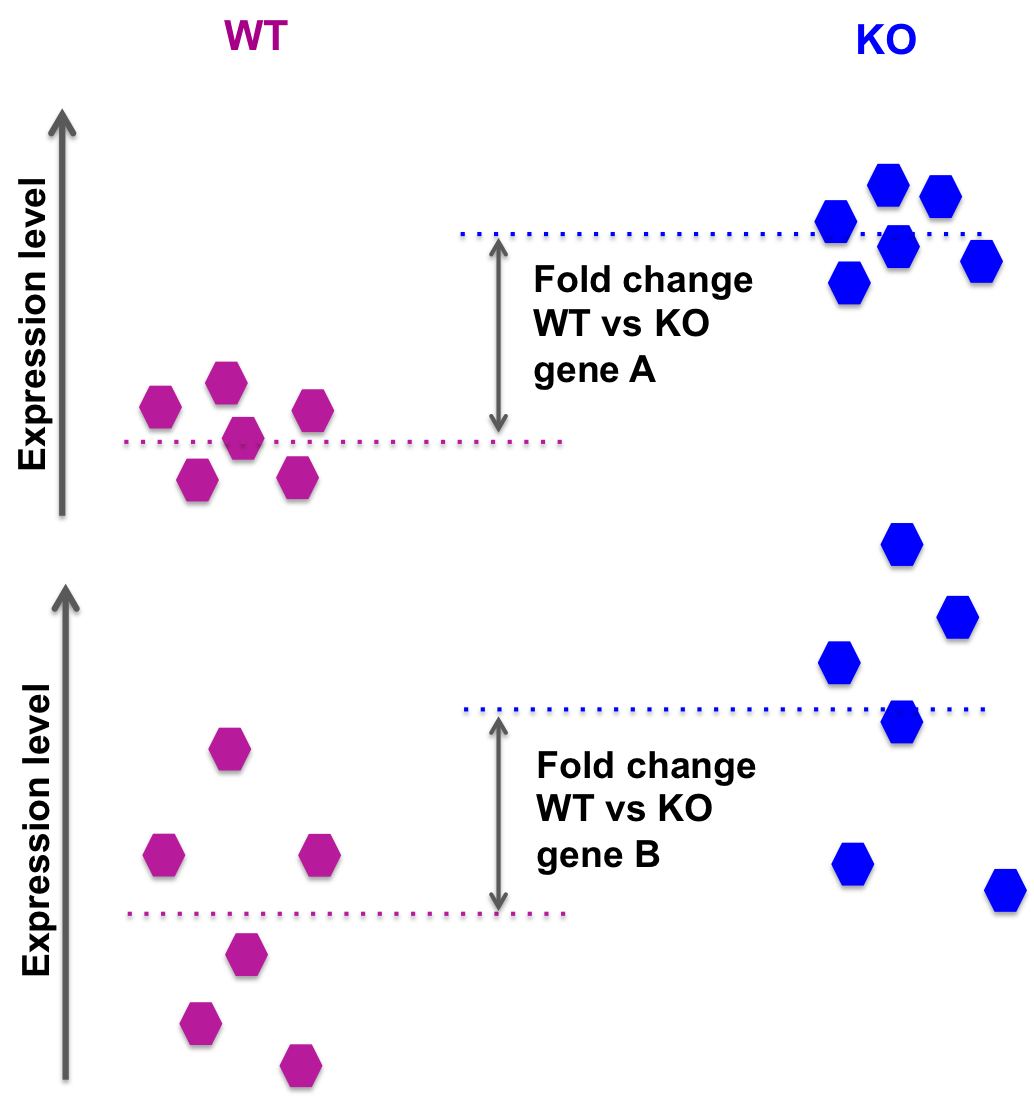
The distribution of log2 fold changes when the expression levels from... | Download Scientific Diagram
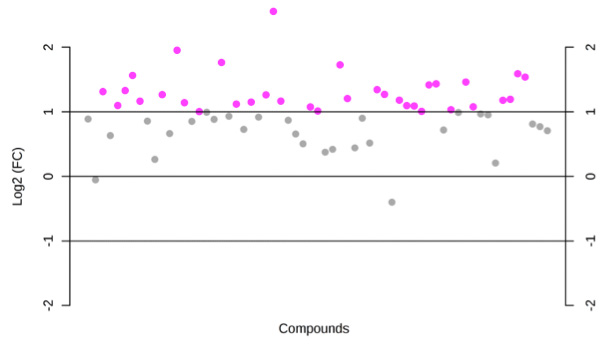
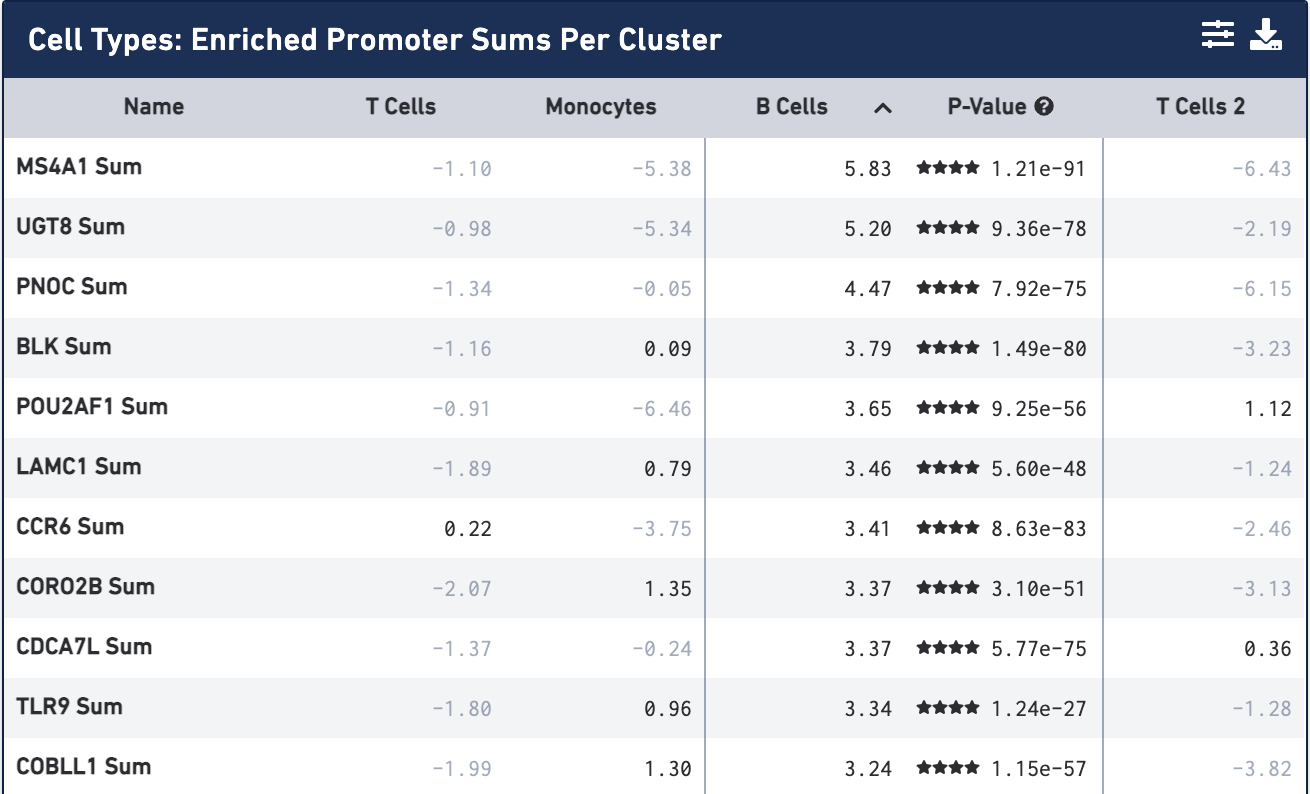
The MicroRNA and MessengerRNA Profile of the RNA-Induced Silencing Complex in Human Primary Astrocyte and Astrocytoma Cells | PLOS ONE