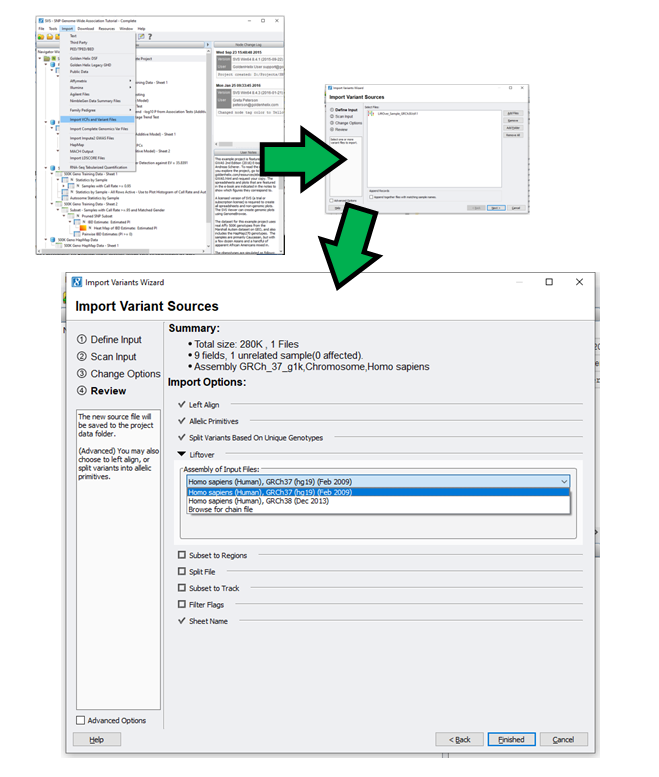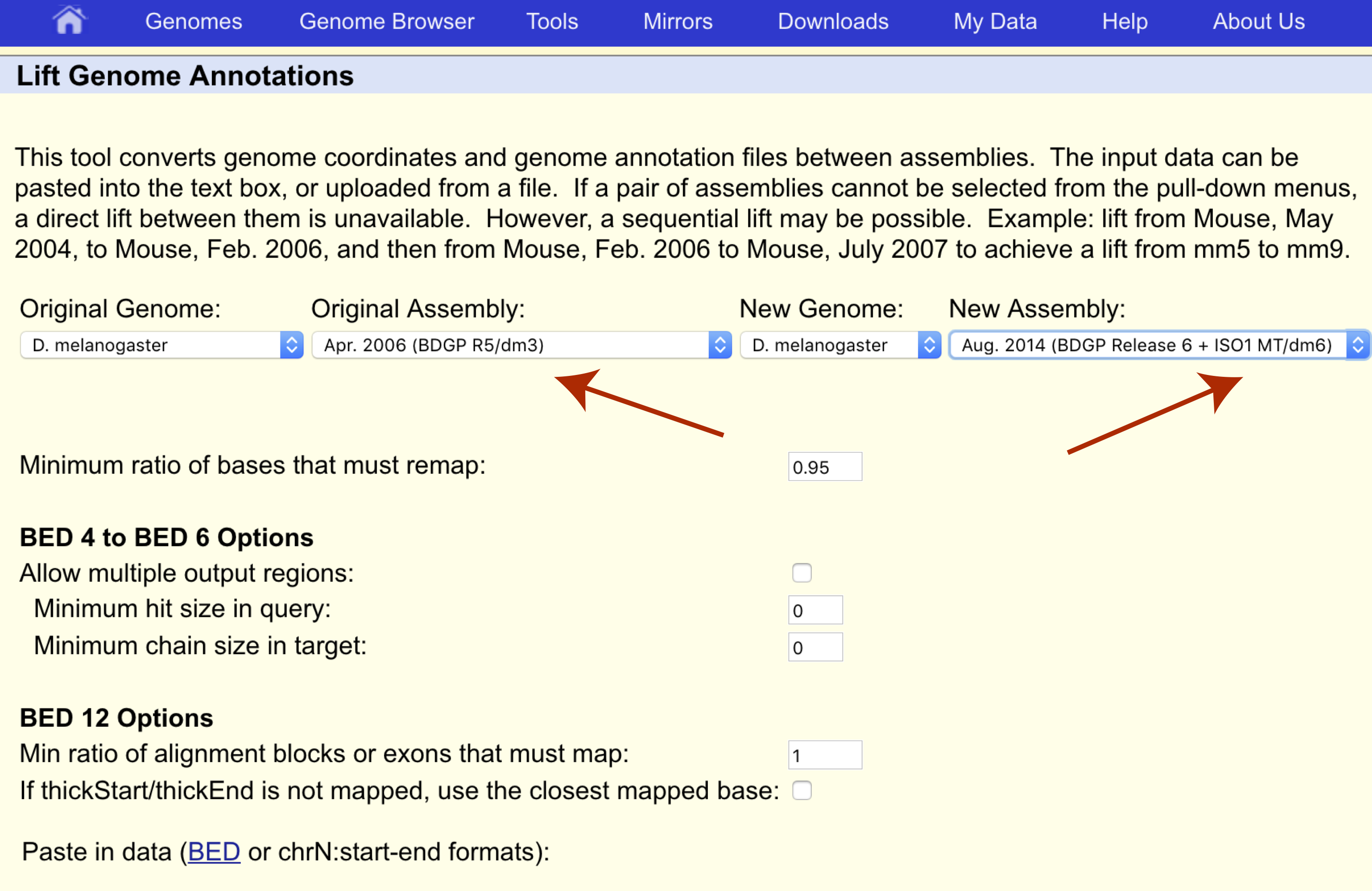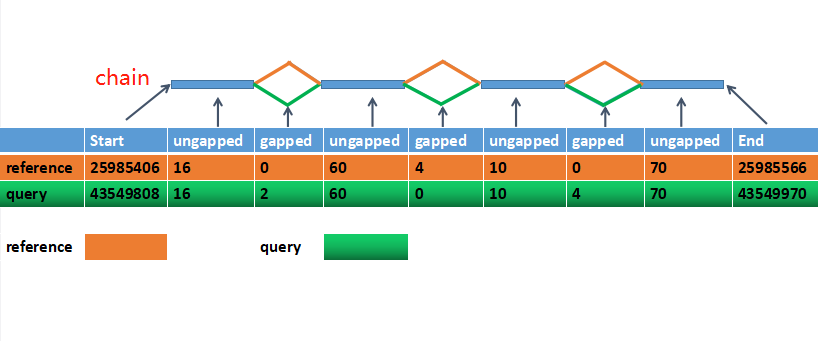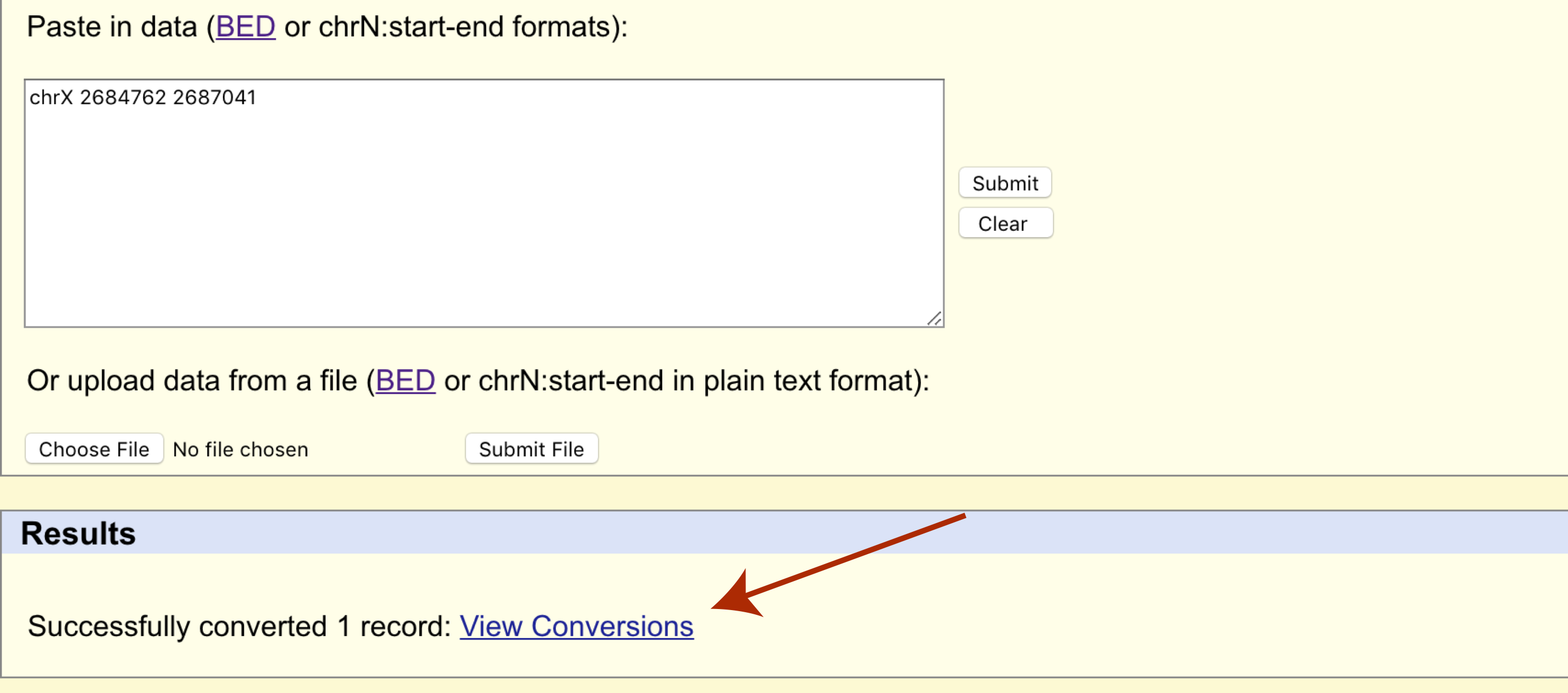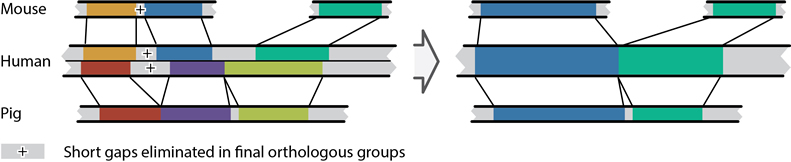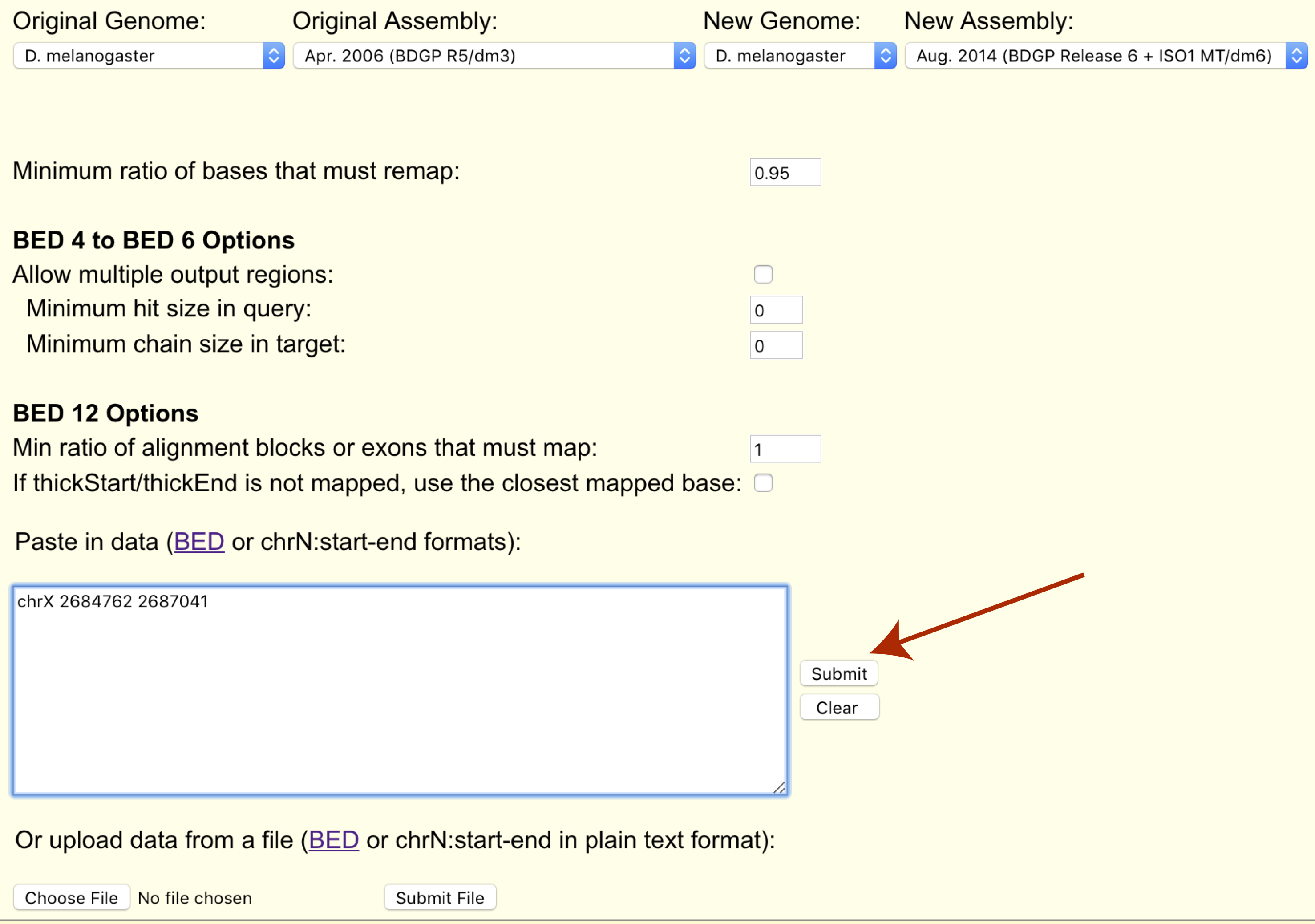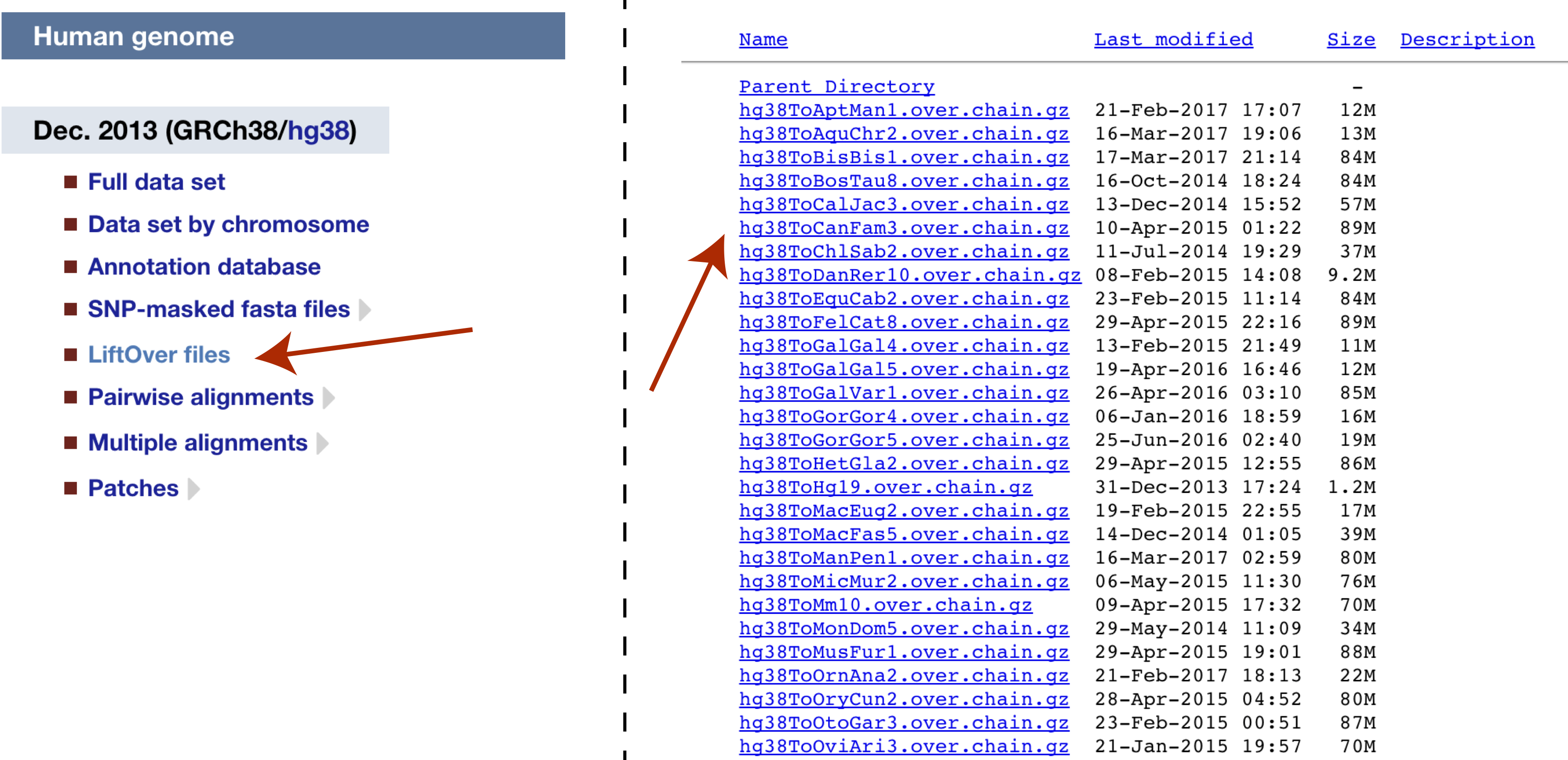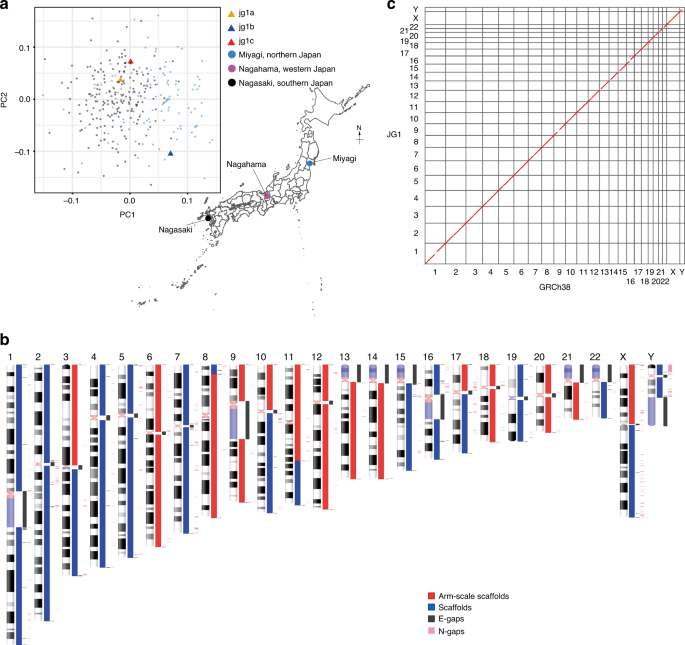
Construction and integration of three de novo Japanese human genome assemblies toward a population-specific reference | Nature Communications

AnchorWave: Sensitive alignment of genomes with high sequence diversity, extensive structural polymorphism, and whole-genome duplication | PNAS

Liftover to Map Genomic Coordinates Between Genome Builds with Rtracklayer Bioconductor Package - YouTube
GitHub - billzt/pyOverChain: a python pipeline to generate chain files between different genome assemblies for LiftOver.

The workflow of segment_liftover: (1) It can take either a folder or a... | Download Scientific Diagram
GitHub - burgshrimps/liftover_T2T: Chain files and scripts for coordinate conversions between hg38 and CHM13 (T2T assembly).
![PDF] Benchmark study comparing liftover tools for genome conversion of epigenome sequencing data | Semantic Scholar PDF] Benchmark study comparing liftover tools for genome conversion of epigenome sequencing data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a939a23b0f9714c27536beb57392d0b03e21e51f/10-Figure4-1.png)